In 1912, on the ancient lava fields of Haleakalā on the Hawaiian island of Maui, a single tree stood near death. Fifteen feet tall, its bark encrusted with lichens, it was down to its last flower.
The Hawaiians called this tree hau kuahiwi, the mountain hibiscus. Unlike the more familiar Hawaiian hibiscus, which grows in moist valleys and opens wide in a welcoming aloha, the mountain hibiscus grew only on the dry, well-drained lava fields of Hawaii’s volcanoes. The plant unfolded only two of its five hibiscuslike petals, keeping the rest closed in a demure, curved tube designed for Maui’s honeycreepers—nectar-eating songbirds with curved bills that were its favored pollinators.
But this tree had not reproduced in years. Most honeycreepers had disappeared as the 19th century gave way to the 20th. The lava fields of Haleakalā had been turned into cattle ranches. Cows rubbed its trunk raw. Rats ate its seeds.
Up the slope came a botanist, dressed in Rough Rider cavalry hat and khakis, a collector’s bag over his shoulder. His name was Gerrit Wilder, and he was on the original expedition that identified this tree in 1910. Because of that, the tree was named for him, Hibiscadelphus wilderianus. It was the only member of its species ever found. Its sickly state was why Wilder had returned. He plucked the last flower, along with some twigs and leaves, and nestled them into his bag. Then he turned and made his way slowly down the slope.
Not long after that, the tree succumbed to the cattle and the rats and dropped its final leaves. H. wilderianus was extinct. And that should have been that. Extinction is supposed to be forever.
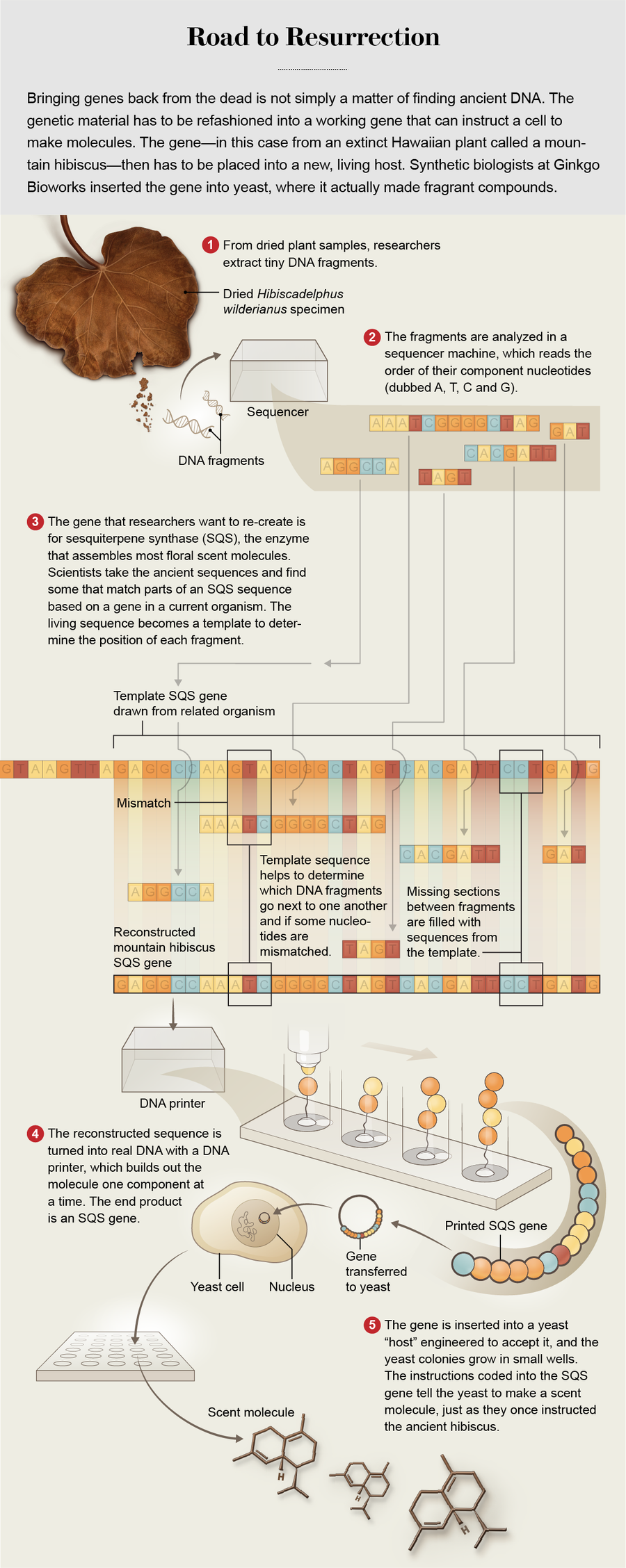
Recent breakthroughs in DNA sequencing, however, have made it increasingly easy to read the genes of long-dead organisms and “reboot” those DNA stretches. Serious efforts are underway to use such tech to revive the passenger pigeon and the woolly mammoth. Both projects depend on bioengineering advances that are still years away. Yet in 2018, in an eighth-floor laboratory built above the burgeoning Seaport District in Boston, a crucial part of this long-dead mountain hibiscus came back to life.
A collection of gene engineers, working for a company called Ginkgo Bioworks, was able to re-create the scent genes from the flower. They rebuilt the genetic material that had produced the flowers’ distinctive odor, got it working again in another life-form—a yeast—and human noses smelled something that had vanished from the planet more than a century ago. Like Odysseus raising the dead in Hades and plying them for information, some kind of communication took place between the living and the deceased. There were no flowers, no petals, but these were the actual DNA sequences of the plant telling cells to churn out molecules as they used to do on Maui, and those molecules were grabbed in people’s noses, sending signals to their brains. It was the most tangible sign yet that the hard membrane of extinction is beginning to soften. This newfound porosity forces a strange question: Can we reboot enough genes to say that something isn’t quite dead anymore?
Scent of life
The resurrection project began, oddly enough, at the 2014 annual convention of the International Federation of Essential Oils and Aroma Trades in Rome, where Jason Kelly, Ginkgo’s CEO, was looking to drum up business. Kelly and his Ginkgo co-founders graduated in 2008 from the Massachusetts Institute of Technology with some of the first Ph.D.s in synthetic biology awarded for that specialization. His firm is also highly specialized: if another company needs a new microbe to produce some valuable molecule—for fuel, fiber, fragrance, pharmaceutical, whatever—Ginkgo will design and test hundreds of prototypes in its biofoundries and hand over the top performers.
Many of Ginkgo’s best clients are in the flavors and fragrances industry, where raw ingredients can be astronomically expensive. All those fragrance molecules are produced by enzymes in the plant cells, and the blueprint for those enzymes is coded in DNA by a gene. Like software, this code can run on any compatible platform, and life is surprisingly platform-agnostic. All living things use the same four-letter language of DNA—components labeled A, T, C and G—and yeast and plants run many of the same genes. By inserting fragrance genes into specially engineered strains of brewer’s yeast, Ginkgo brews scent molecules in a flask, just like making beer.
At the trade show, Kelly met a consultant for Givaudan, the Swiss perfume giant, who told Kelly about Givaudan’s Scent Trek program, which dispatched explorers into the world’s rain forests to capture the air around rare flowers so the scents could be identified. Kelly was intrigued. If Ginkgo could get samples of these plants, the company could sequence the genes and synthesize the enzymes that made the smells. But as the two brainstormed, Kelly had a much crazier idea. What if he was able to go beyond obscure plants and bring back the scent of flowers that no longer even existed?

This would be the first step, he thought, in reversing a tremendous biological waste. “The planet has spent three billion years trying out different DNA sequences through this process we call evolution,” Kelly says, “and that’s what we have today. But along the way, a lot got lost for some random reason—a meteor or whatever—and some of that stuff was incredible. The planet spent hundreds of millions of years evolving DNA. And we just have to let it go away? For a biological designer, it’s frustrating to imagine losing all that great code.”
Kelly’s original plan riffed on Jurassic Park: Recover an Ice Age flower from the Arctic permafrost, sequence its genes and synthesize the ones responsible for fragrance, then put them in yeast cells. When the genes instructed the cells to make the fragrant molecules, Kelly could brew up a little Extinction No 5.
It was a long shot. Although a handful of ancient genes had been reconstructed in labs, most simply sat there, never being asked to produce a protein and thus rejoin our world. Even if Ginkgo was able to rebuild old genes, those genes might not function in new yeast. Kelly also worried about tying up precious resources. Everyone at his company was already overworked. The last thing they needed was to get sucked into a Jurassic lark.
But the project found a champion in Christina Agapakis, Ginkgo’s creative director. Agapakis earned her Ph.D. in synthetic biology at Harvard University, and she worked on optimizing bacteria to produce hydrogen fuel and created art based on the shapes of antibodies. Warm and witty, she was drawn to research that probed the borderlands between natural and unnatural and opened up interesting conversations about genetically modified organisms. A perfume of extinct florals that people could smell while meditating on these lost species was right up her alley. She dubbed the venture “Project Cretaceous,” after the period when flowers first came into existence. She began by contacting experts in Ice Age excavations, who told her it was impossible to sequence a full genome out of the gunky specks of plant that emerge from the permafrost. The Ice Age was a dead end.
Before giving up, Agapakis did what any good Millennial would do: she googled “extinct plant DNA sequencing.” Far down the list of results, she found an obscure paper from the Biological Journal of the Linnean Society on museomics, a new technique for extracting DNA from museum-preserved plants and animals. So she did not need permafrost after all. She just needed an herbarium.
The realization made the Harvard grad smile. She knew just where to find one of those.
The DNA search
The Harvard Herbaria, which date back to 1842, anchor one brick-lined end of a street named Divinity Avenue, and their numerous floors are filled with formaldehyde-scented cabinets holding more than five million samples. They do not embrace change enthusiastically, so when Agapakis pitched her plan in 2016, the curator was skeptical. Do what with their plants? The herbaria were not in the business of giving away their collection to for-profit entities. Besides, they had no searchable database for their holdings, so they had no idea if they had any extinct plants or not.
It took Agapakis months of negotiations to reach an agreement. The deal was sealed when she offered to provide genomes of any extinct plants she found to the research community. Even so, she had to find the plants on her own, with no help from herbaria staff, and if she did find what she was looking for she could not take more than a pinky-nail-sized fragment of extraneous material.
Agapakis and Dawn Thompson, Ginkgo’s head of Next Generation Sequencing, printed out the IUCN Red List of 116 modern plant extinctions and began their quest. The collection was arranged by plant family first and geography second, so the only way to find a sample was to go to the corresponding floor of the herbaria, find the aisle for the right family and then search in all the folders for the particular country or area. The aisles were endless, the cabinets seemingly filled with everything but the plants they were looking for. Then, in the Hawaii room, Agapakis cranked a big wheel to roll the creaking cabinets apart, opened the doors, paged through the folders, opened one, and gazed down at three long twigs holding an array of broad, beautiful leaves and a single pressed flower bud. “Flora of the Hawaiian Islands” read the attached card. “Hibiscadelphus wilderianus.” Agapakis felt an electric thrill. It was Wilder’s extinct tree, right in front of her.
In the end, the scientists found 20 of the plants on their list in the herbaria, 14 of which had enough material to spare. Under the baleful eye of the curator, they snapped off the least important bits and placed them in plastic baggies.
Then it was time for the hard part. DNA degrades after an organism dies. Ginkgo was going to have to find needles of DNA in haystacks of cellulose. And the team had only enough material for a few attempts. The researchers decided to practice on an oak leaf scavenged from the streets of Boston. Even that did not go well. Despite their state-of-the-art sequencing equipment, they struggled to extract DNA from the samples. The ancient samples did not produce anything.
With pressure mounting to yield the sequencing machine to paying projects, Agapakis and Thompson had a sobering conversation. If they kept trying, they were going to run out of plant material, and there was no way they were getting more. They decided to put Project Cretaceous on hold until they found a more effective way of doing it.
Months later, at a conference, Kelly met Beth Shapiro, co-director of the Paleogenomics Lab at the University of California, Santa Cruz. This is the place you go if you want to de-extinct a mammoth or a passenger pigeon. Every year it gets better and better at extracting tiny amounts of DNA from iffy old material. In 2016 the lab was able to identify 0.01 to 0.05 percent mammoth DNA—a mere whiff of pachyderm—in 5,650-year-old lake sediments from an island in the Bering Strait. Send us your flowers, Shapiro said.
Thompson overnighted her plastic baggies of leaf matter to Josh Kapp, a grad student in the Paleogenomics Lab. Kapp did not like what he saw. He pulverized each sample into powder to maximize the surface area, but the plants did not powder as nicely as the bone he was used to. But after many filtration steps and some creative applications of chemicals that bind to DNA fragments, Kapp ended up with 14 microtubes holding the secrets of lost plants, which he packed in dry ice and sent back to Ginkgo. When Thompson ran the samples through her sequencing machine in Boston, she was thrilled to see numerous short reads come through: millions of fragments of genetic code, each just 40 or 50 letters long.

Reconstruction in action
But did any of those fragments belong to a scent gene, and could they be put back together? Ginkgo was looking for genes, typically about 1,700 letters long, that made enzymes called sesquiterpene synthases (SQSs); these are the enzymes that stitch together most good floral scent molecules. A typical flower might have several of these genes. With all the tiny fragments they had recovered, it was as if the Ginkgo researchers had a book for each plant—the extinct plant genome—all chopped up into random 50-letter chunks and mixed together, and they needed to reassemble a few 1,700-letter passages in just the right way.
If the scientists had copies of the original books to use as a template or even a few chapters, they could figure out where the fragments went. Here evolution came to the rescue. It never invents anything from scratch. New species evolve from older species, tweaking or repurposing the original genes. So most SQS genes in modern plants share a lot of DNA code with closely related ancestors. Jue Wang, a computational biologist then working at Ginkgo, was tasked with this book-reassembly problem. He realized those modern SQSs could serve as the template. It was like trying to reconstruct a lost version of the Bible using the King James and New International versions as guides. The wording would not exactly match, but they would be a decent guide to what went where.
Bit by bit, Wang built his genes on the scaffolding modern relatives provided, relying on sequence overlaps for placement. He filled in any missing letters of DNA from the modern templates. If his fragmented Bible read “In th_ beg_ _ning was the W_ _ d,” he could look to the King James and be pretty confident which letters were missing.
Ultimately Wang was able to reconstruct 2,738 versions of genes from the extinct flowers. Undoubtedly these strings of biological letters had a few typos. Would that ruin their functions? Occasionally a single wrong letter of DNA will break a gene catastrophically, as in sickle cell anemia. But often minor changes do not affect the end product. In fact, sometimes genes with significantly different forms will function similarly. In Biblical terms, “In the beginning was the Word” (King James) and “The Word was first” (The Message) do not match letter by letter, but both get the job done. Wang thought most of his letter strings would be too glitchy. He just hoped a few would work as instructions for a real cell.
For that to happen, these genes, which existed solely in Wang’s computer, had to be converted into physical DNA. That is a fairly straightforward job, done with a DNA printer that resembles a 3-D printer but shoots out As, Cs, Gs and Ts, which bind together chemically into a classic double helix. Although this is often called synthetic DNA, it is just as real as any other DNA. Molecules are molecules.
Then it was up to the yeast, each of the 2,000-odd genes going into a colony bred to accept new DNA and make molecules according to its instructions. For several days the colonies frothed like beer brewing in their tiny containers. Scott Marr, a molecular microbiologist at Ginkgo, watched, wondering what they had made. When the fermentation subsided, Marr ran a sample of each colony through a mass spectrometer, a kind of artificial nose that was capable of detecting and identifying the minuscule amount of molecules being produced in each strain. Each mass shows up as a differently sized peak on a graph. It was Marr’s job to read the pattern of peaks like a fingerprint.
He wrote programs to eliminate all the regular products of yeast metabolism in the machine’s readout, so only nonyeast SQS products—scent-making sesquiterpenes—would show up. Mindful of the long odds and the likelihood of typos in the Ginkgo translations, Marr crossed his fingers and ran the samples. The readouts showed nothing. Then more nothing. It looked like the scientists had pieced together book passages with too many letter mistakes, paragraphs that no cell could read.
And then there it was: a peak. After a while, there was another and another. Marr let out a pent-up breath and began to match the molecular fingerprints to his database of terpenes. Then he broke the good news to the Project Cretaceous team: dozens of the flower-yeast chimeras were alive.
Agapakis sat at a table, listening to Marr’s report and taking it all in. It had been three years since the initial crazy idea. Many times she and her colleagues had nearly abandoned it. And now they had molecules. Real molecules! Made by genes that had not existed in a century!

Back in the real world
Ginkgo’s yeast was able to get genes from three different extinct plants to produce sesquiterpenes. Although the microscopic amounts were far too small to smell directly, the scientists had some inklings of what the eventual floral nature might be, based on the smells of modern counterparts. One of the plants, the Falls-of-the-Ohio scurfpea—a legume that made the fatal mistake of growing only on a few rocky islets in the Ohio River that were drowned by dams in the 1920s—produced a handful of sesquiterpenes that, if some 21st-century relatives were any guide, would have woody, peppery, balsamic scents.
The Wynberg conebush—a five-foot-tall flower with white petals and a yellow head that grew in the granite hills above Cape Town until 1806, when it disappeared forever underneath South Africa’s expanding vineyards—produced an astonishing 21 sesquiterpenes, many of which are associated with tantalizing scents: jasmine, lemongrass, cannabis, chamomile, turmeric, ginger, hops. That awkward mix sounded like a good match for a flower that had been noted for its “strong and disagreeable smell.”
Eleven sesquiterpenes came from H. wilderianus, the mountain hibiscus, which had last released its essence to the world in 1912, as Gerrit Wilder picked that final flower and descended Haleakalā, never expecting that anyone would smell the hau kuahiwi again. From there, the genes’ unlikely journey along Resurrection Road had taken them to the College of Hawaii’s herbarium, where the plant was dried and pressed and eventually shared with the Harvard Herbaria. There it waited for decades for Agapakis to open its manila folder and break off a piece of the corpse. The genes were liquefied in Santa Cruz, digitized in Boston, then reanimated in the tender embrace of an organism completely unlike the one that hosted their last appearance on planet Earth. The genes had crossed time and space and outward form, but their information held.
And then it was time to smell them. The Project Cretaceous team picked the Hibiscadelphus to go first because its allure was captivating, much as it had been for many a honeycreeper for millennia. On a bright August day in a crisp white conference room, the group gathered to sample a variety of formulations—created for the company by Berlin-based scent artist Sissel Tolaas—that blended the Hawaiian molecules in different combinations and concentrations. One of the molecules, juniper camphor, was a pricey ingredient in fragrant oils. Hibiscadelphus had expensive tastes.
They dipped paper fragrance test strips into 11 elfin bottles, held them a few inches from their noses and sniffed gently. Team members grinned at one another as if they could not quite believe they were here. “First resurrected fragrance!” Kelly announced. Agapakis’s reaction was more visceral. “I feel overwhelmed,” she said. “I couldn’t imagine what this was going to smell like.”
Some samples had flashes of citrus or thyme. All had a woody core of bark and juniper that must have been the essence of hau kuahiwi. “I like the lightness,” Agapakis said, eyes closed as she inhaled. “It feels ethereal.”
Lurking in the background of several samples was a smoky hint of sulfurous dirt. Kelly’s eyes twinkled as he held one under his nose. “This is pretty magical, to be honest,” he said. “I hope it captures people’s imagination and gets them to think about what we’ve lost.”
The scent—and the thoughts it inspires—is an important milestone, says Stanford University bioengineer Megan Palmer, a board member of Revive & Restore, a nonprofit that is supporting the passenger pigeon and woolly mammoth resurrection projects. “We can’t know exactly what these flowers smelled like,” she says, “but we can get molecular hints that we interpret through what we know about the species we see in the world today.” As scientific advances, she adds, “these techniques can help us make smarter guesses at how extinct species functioned. They may even allow more ambitious projects to restore those functions and the species that gave rise to them.”
Because of this work, we are a tiny bit closer to coaxing saber-toothed tiger musk or Neandertal hemoglobin out of cells. And as more of these freelance genes return to function in new forms, they make us begin to question our old emphasis on species. The traditional genetic container may not limit the life of its contents. Sitting in that Boston conference room, it seemed clear that one of the most opportune moments in DNA’s four-billion-year career had begun. This novel environment of bioengineering labs and digital databases and DNA printers was giving genes a newfound freedom to flow, new ways to replicate, new habitats to populate, new organisms to seduce. The original form may go extinct, but many functions can return, and at some point—nobody really knows what that is—that resurrection may get an organism to the point of “no longer dead.”
As the essential oils saturated the air, the room became an unlikely tropical oasis, a hint of smoke in the distance, and it was easy to imagine the sun-baked lava fields of Haleakalā in the ancient past, a forest of mountain hibiscus all around, bright red honeycreepers flitting from blossom to blossom. That world will never come again, but some of the countless genes from primordial Hawaii and other lost landscapes may do just that. They are pressing against the membrane of extinction at this very moment, probing, hungry for any chance to get back in the action.